QC sample correlation plot
Xiaotao Shen (https://www.shenxt.info/)
Created on 2021-12-04 and updated on 2022-09-19
Source:vignettes/massqc_qc_correlation.Rmd
massqc_qc_correlation.RmdData preparation
library(massdataset)
library(tidyverse)
library(massqc)
data("sample_info", package = "massdataset")
data("expression_data", package = "massdataset")
data("variable_info", package = "massdataset")
object =
create_mass_dataset(
expression_data = expression_data,
sample_info = sample_info,
variable_info = variable_info
)
object
#> --------------------
#> massdataset version: 0.99.1
#> --------------------
#> 1.expression_data:[ 1000 x 8 data.frame]
#> 2.sample_info:[ 8 x 4 data.frame]
#> 3.variable_info:[ 1000 x 3 data.frame]
#> 4.sample_info_note:[ 4 x 2 data.frame]
#> 5.variable_info_note:[ 3 x 2 data.frame]
#> 6.ms2_data:[ 0 variables x 0 MS2 spectra]
#> --------------------
#> Processing information (extract_process_info())
#> create_mass_dataset ----------
#> Package Function.used Time
#> 1 massdataset create_mass_dataset() 2022-01-16 22:54:43QC correlation plot
object %>%
massqc_sample_correlation()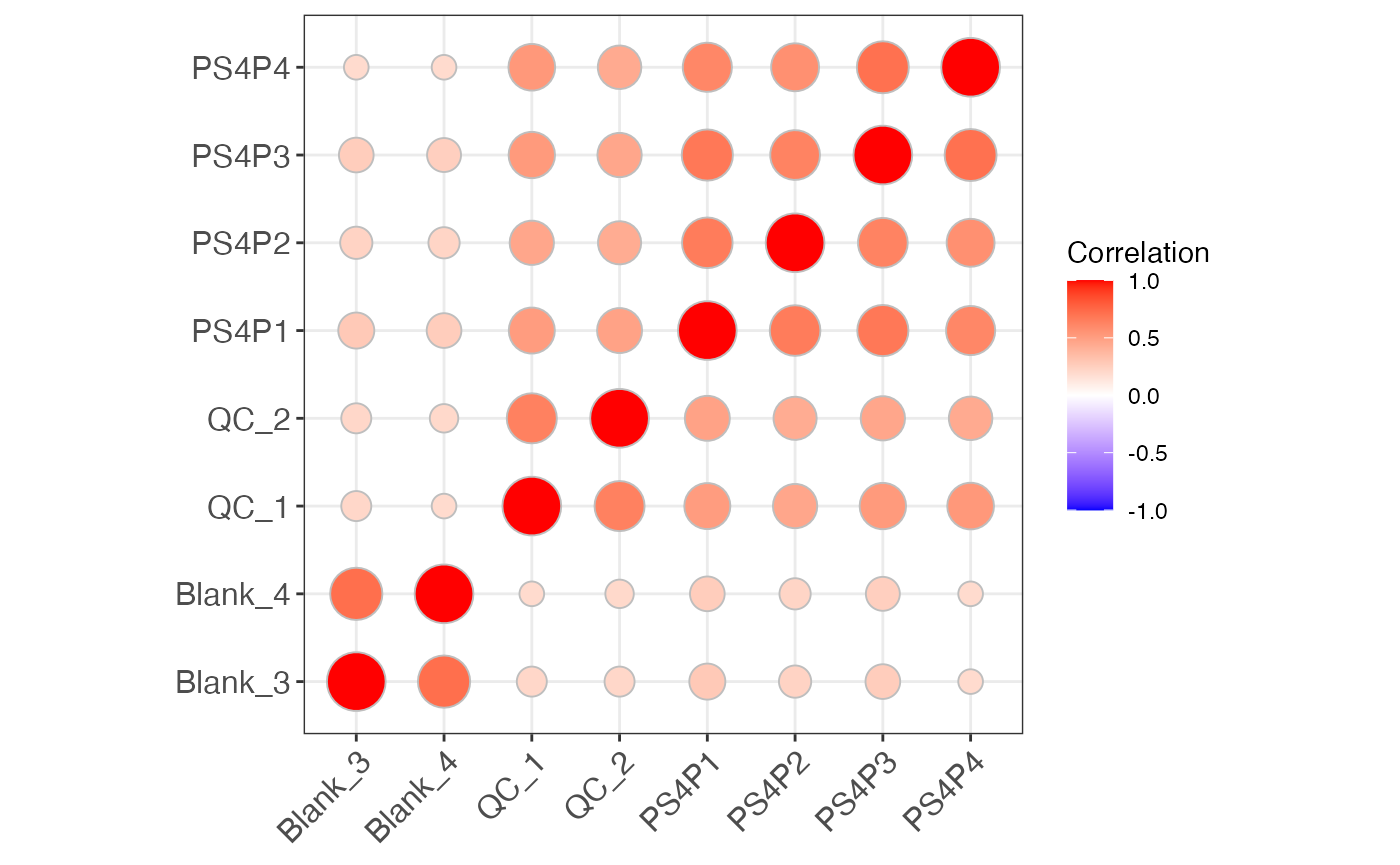
object %>%
massqc_sample_correlation()
object %>%
activate_mass_dataset(what = "sample_info") %>%
filter(class == "QC") %>%
massqc_sample_correlation()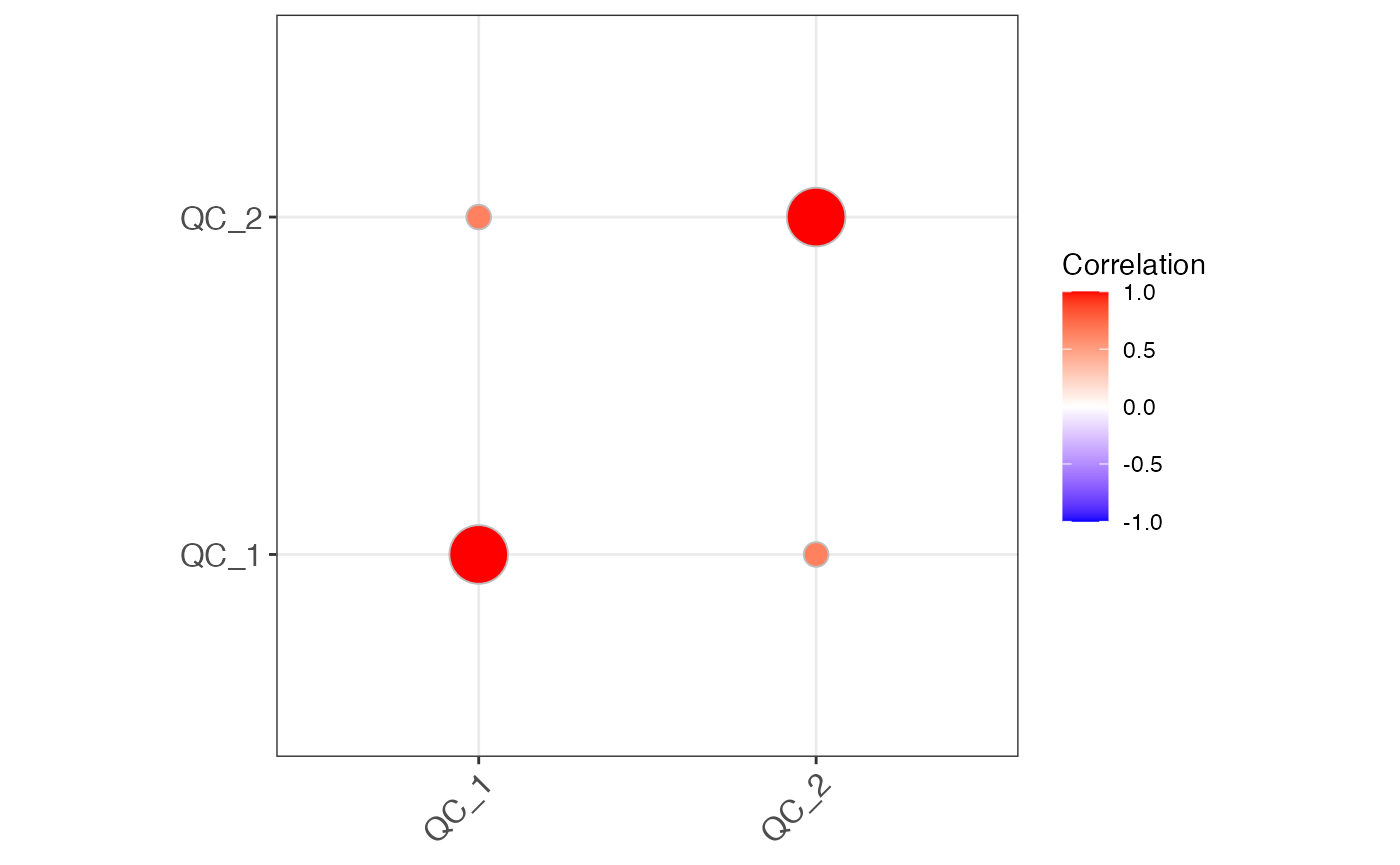
object %>%
massqc_sample_correlation(lab = TRUE)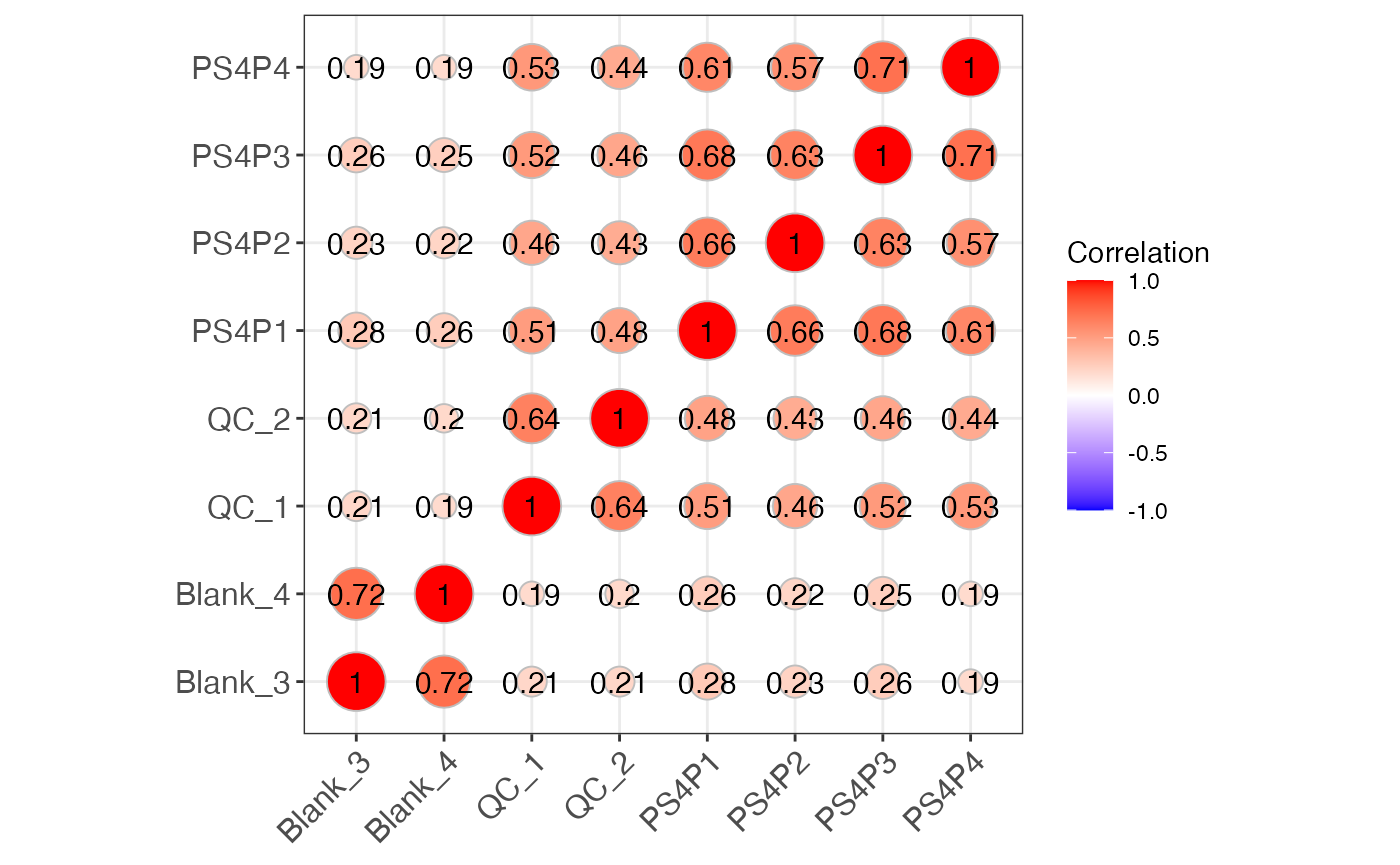
object %>%
massqc_sample_correlation(lab = TRUE, type = "upper")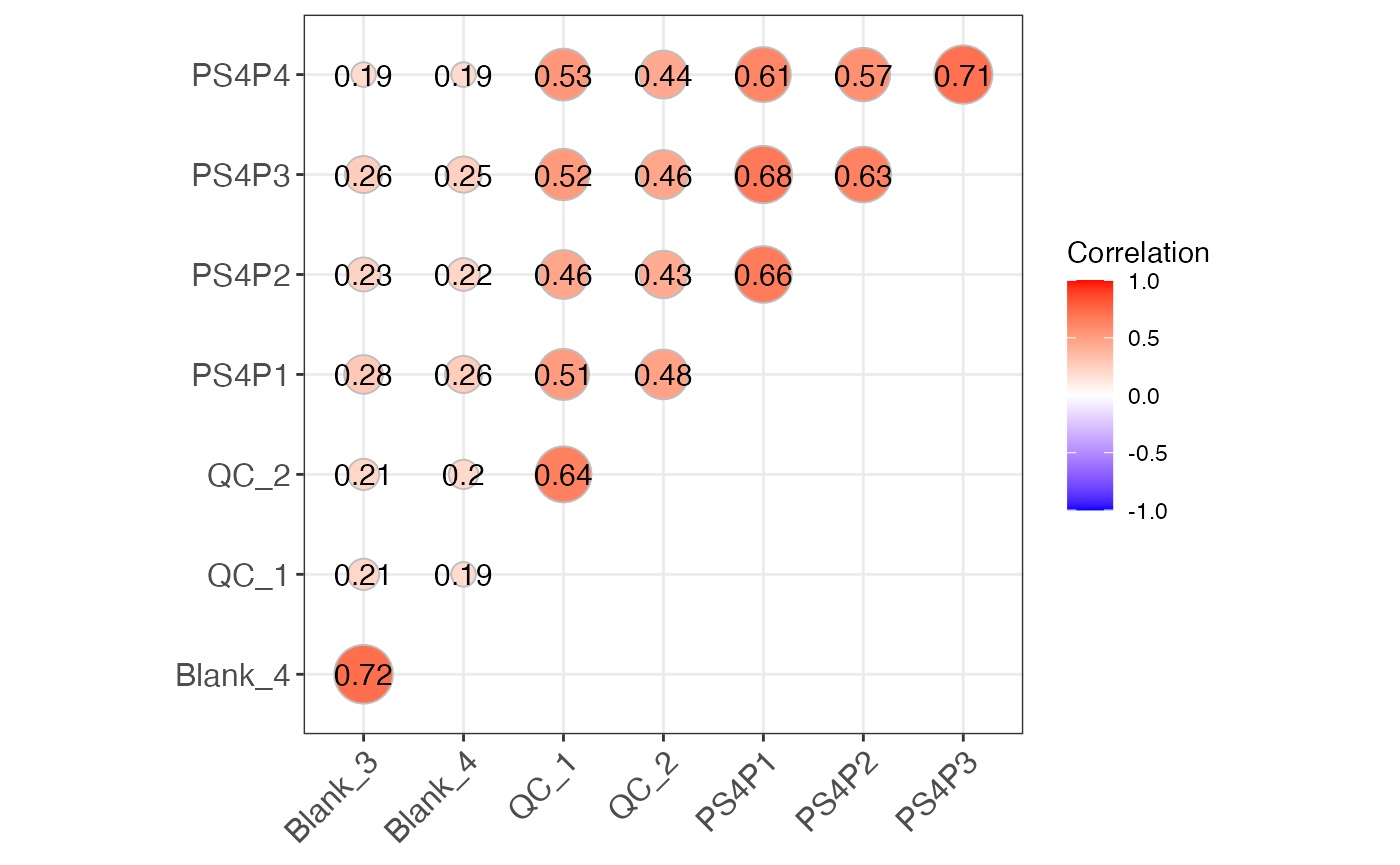
Session information
sessionInfo()
#> R Under development (unstable) (2022-01-11 r81473)
#> Platform: x86_64-apple-darwin17.0 (64-bit)
#> Running under: macOS Big Sur/Monterey 10.16
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggfortify_0.4.14 massqc_0.0.1 forcats_0.5.1 stringr_1.4.0
#> [5] dplyr_1.0.7 purrr_0.3.4 readr_2.1.1 tidyr_1.1.4
#> [9] tibble_3.1.6 ggplot2_3.3.5 tidyverse_1.3.1 magrittr_2.0.1
#> [13] tinytools_0.9.1 massdataset_0.99.1
#>
#> loaded via a namespace (and not attached):
#> [1] colorspace_2.0-2 rjson_0.2.21 ellipsis_0.3.2
#> [4] leaflet_2.0.4.1 rprojroot_2.0.2 circlize_0.4.14
#> [7] GlobalOptions_0.1.2 fs_1.5.2 clue_0.3-60
#> [10] rstudioapi_0.13 farver_2.1.0 ggrepel_0.9.1
#> [13] fansi_1.0.0 lubridate_1.8.0 xml2_1.3.3
#> [16] codetools_0.2-18 doParallel_1.0.16 cachem_1.0.6
#> [19] knitr_1.37 jsonlite_1.7.2 broom_0.7.11
#> [22] cluster_2.1.2 dbplyr_2.1.1 png_0.1-7
#> [25] compiler_4.2.0 httr_1.4.2 backports_1.4.1
#> [28] ggcorrplot_0.1.3 assertthat_0.2.1 fastmap_1.1.0
#> [31] lazyeval_0.2.2 cli_3.1.0 htmltools_0.5.2
#> [34] tools_4.2.0 gtable_0.3.0 glue_1.6.0
#> [37] reshape2_1.4.4 Rcpp_1.0.7 Biobase_2.55.0
#> [40] cellranger_1.1.0 jquerylib_0.1.4 pkgdown_2.0.1
#> [43] vctrs_0.3.8 iterators_1.0.13 crosstalk_1.2.0
#> [46] xfun_0.29 openxlsx_4.2.5 rvest_1.0.2
#> [49] lifecycle_1.0.1 scales_1.1.1 ragg_1.2.1
#> [52] clisymbols_1.2.0 hms_1.1.1 parallel_4.2.0
#> [55] RColorBrewer_1.1-2 ComplexHeatmap_2.11.0 yaml_2.2.1
#> [58] memoise_2.0.1 pbapply_1.5-0 gridExtra_2.3
#> [61] pander_0.6.4 yulab.utils_0.0.4 sass_0.4.0
#> [64] stringi_1.7.6 highr_0.9 S4Vectors_0.33.10
#> [67] desc_1.4.0 foreach_1.5.1 BiocGenerics_0.41.2
#> [70] zip_2.2.0 shape_1.4.6 rlang_0.4.12
#> [73] pkgconfig_2.0.3 systemfonts_1.0.3 matrixStats_0.61.0
#> [76] evaluate_0.14 labeling_0.4.2 patchwork_1.1.1
#> [79] htmlwidgets_1.5.4 tidyselect_1.1.1 ggsci_2.9
#> [82] plyr_1.8.6 R6_2.5.1 IRanges_2.29.1
#> [85] generics_0.1.1 DBI_1.1.2 pillar_1.6.4
#> [88] haven_2.4.3 withr_2.4.3 modelr_0.1.8
#> [91] crayon_1.4.2 utf8_1.2.2 plotly_4.10.0
#> [94] tzdb_0.2.0 rmarkdown_2.11 GetoptLong_1.0.5
#> [97] grid_4.2.0 readxl_1.3.1 data.table_1.14.2
#> [100] reprex_2.0.1 digest_0.6.29 gridGraphics_0.5-1
#> [103] textshaping_0.3.6 stats4_4.2.0 munsell_0.5.0
#> [106] viridisLite_0.4.0 ggplotify_0.1.0 bslib_0.3.1