Boxplot for each samples.
massqc_pca(
object,
color_by,
point_alpha = 0.8,
frame = TRUE,
frame.type = "norm",
line = TRUE,
...
)Arguments
- object
tidymass-class object.
- color_by
which column (sample_info) is used to color samples
- point_alpha
point_alpha
- frame
?ggplot2::autoplot
- frame.type
?ggplot2::autoplot
- line
Add lines or not.
- ...
other paramters for ggplot2::autoplot
Value
ggplot2 plot.
Examples
library(massdataset)
library(ggplot2)
data("expression_data")
data("sample_info")
data("variable_info")
object =
create_mass_dataset(
expression_data = expression_data,
sample_info = sample_info,
variable_info = variable_info
)
object %>%
massqc_pca()
#> Warning: MVs in you object,
#>
#> will remove variables > 50% and imputate with zero.
#> Warning: no scale for this dataset, try to scale() before pca.
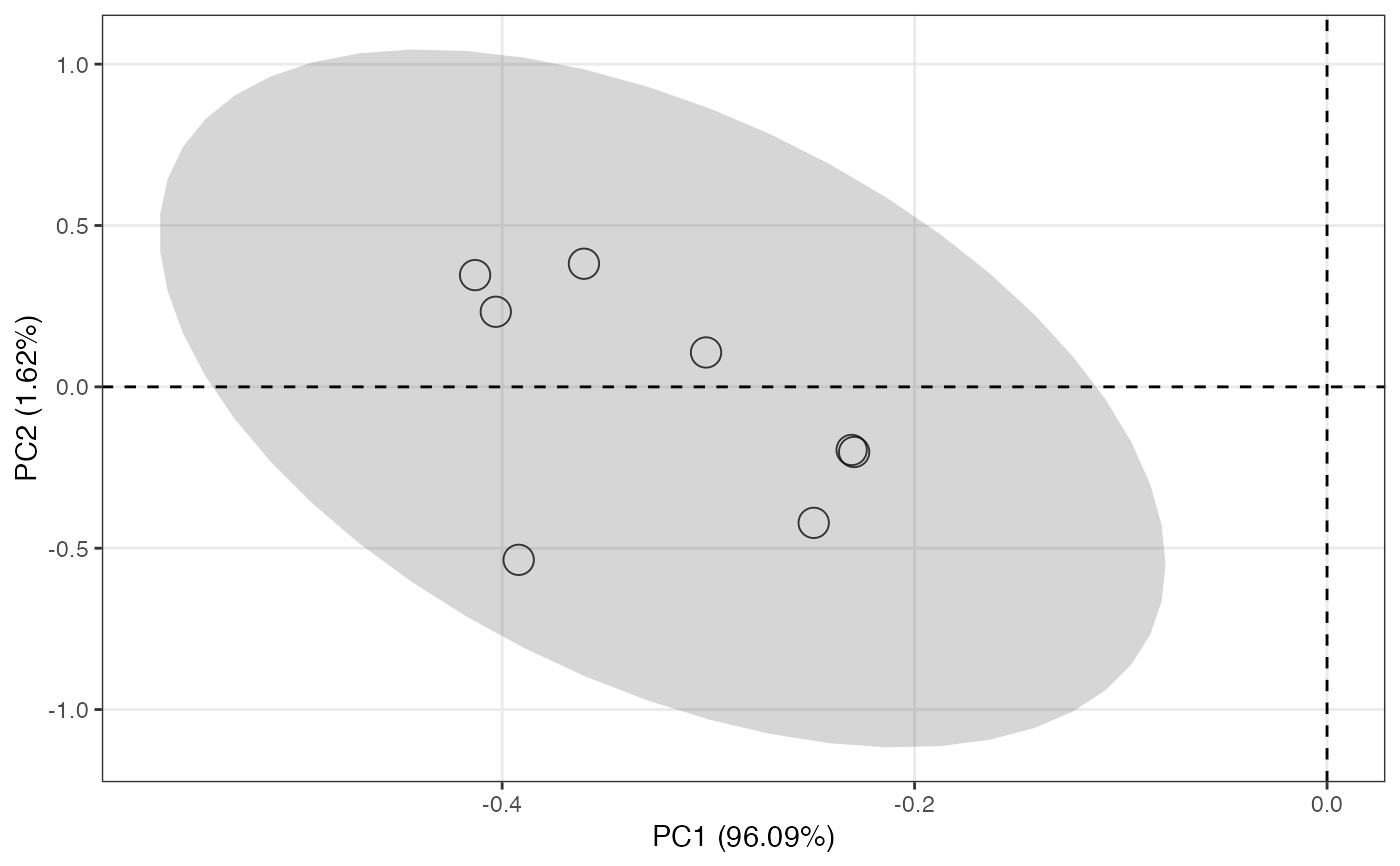 object %>%
massqc_pca(color_by = "class")
#> Warning: MVs in you object,
#>
#> will remove variables > 50% and imputate with zero.
#> Warning: no scale for this dataset, try to scale() before pca.
#> Too few points to calculate an ellipse
#> Too few points to calculate an ellipse
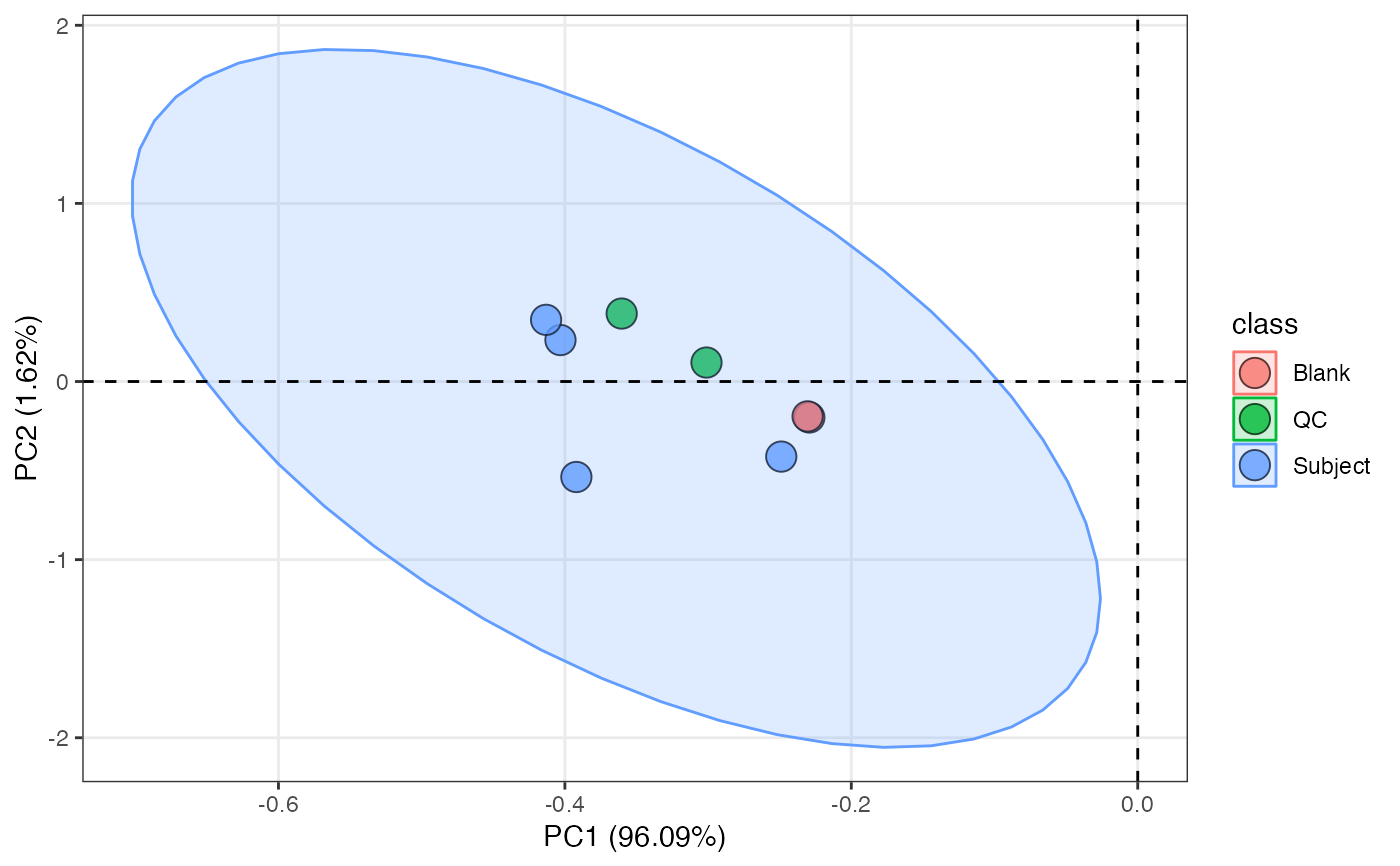
object %>%
massqc_pca(color_by = "class")
#> Warning: MVs in you object,
#>
#> will remove variables > 50% and imputate with zero.
#> Warning: no scale for this dataset, try to scale() before pca.
#> Too few points to calculate an ellipse
#> Too few points to calculate an ellipse
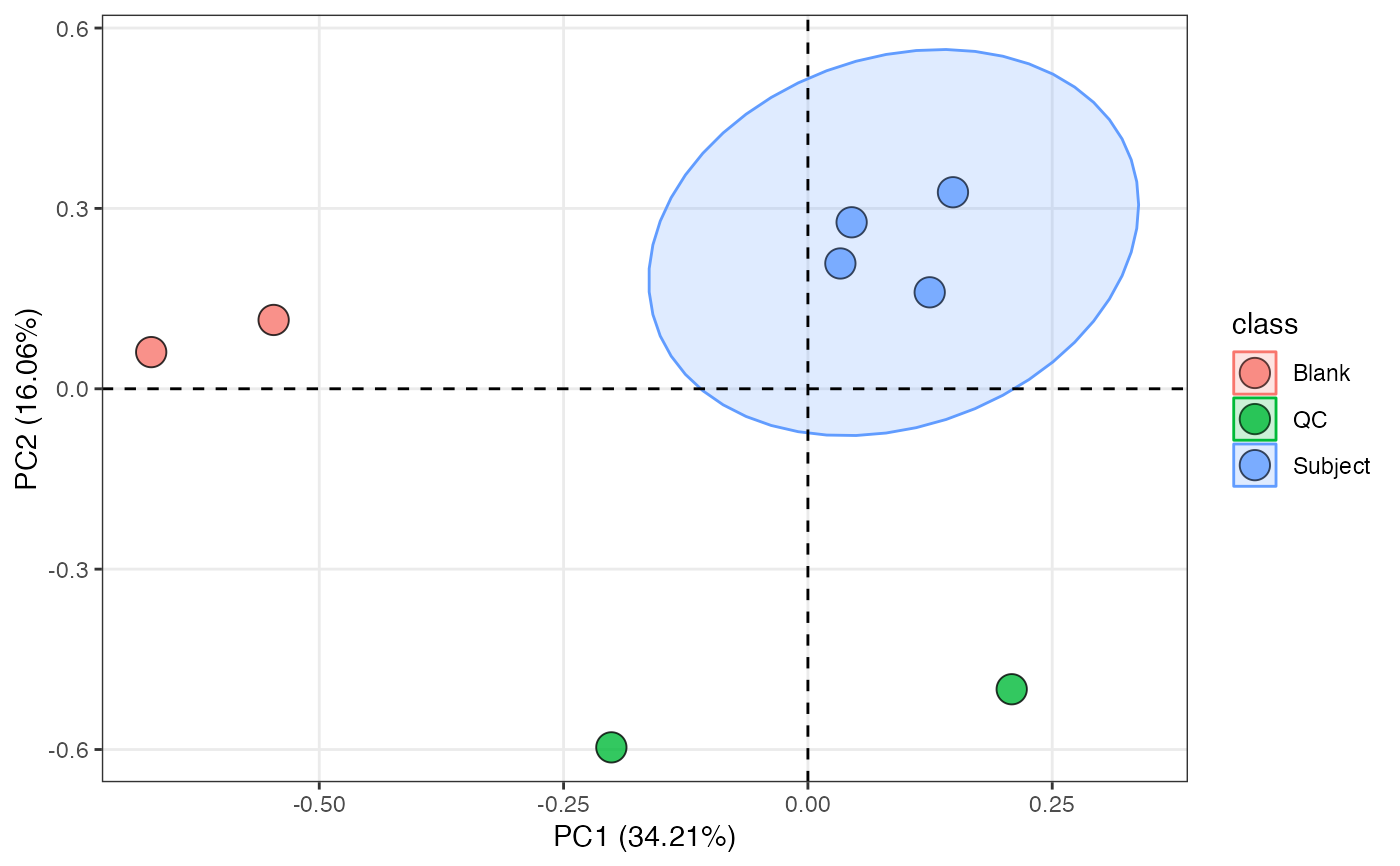
 object %>%
scale %>%
massqc_pca(color_by = "class")
#> Warning: MVs in you object,
#>
#> will remove variables > 50% and imputate with zero.
#> Too few points to calculate an ellipse
#> Too few points to calculate an ellipse
object %>%
scale %>%
massqc_pca(color_by = "class")
#> Warning: MVs in you object,
#>
#> will remove variables > 50% and imputate with zero.
#> Too few points to calculate an ellipse
#> Too few points to calculate an ellipse
 object %>%
scale %>%
massqc_pca(color_by = "class", frame = FALSE) +
ggsci::scale_fill_lancet()
#> Warning: MVs in you object,
#>
#> will remove variables > 50% and imputate with zero.
object %>%
scale %>%
massqc_pca(color_by = "class", frame = FALSE) +
ggsci::scale_fill_lancet()
#> Warning: MVs in you object,
#>
#> will remove variables > 50% and imputate with zero.
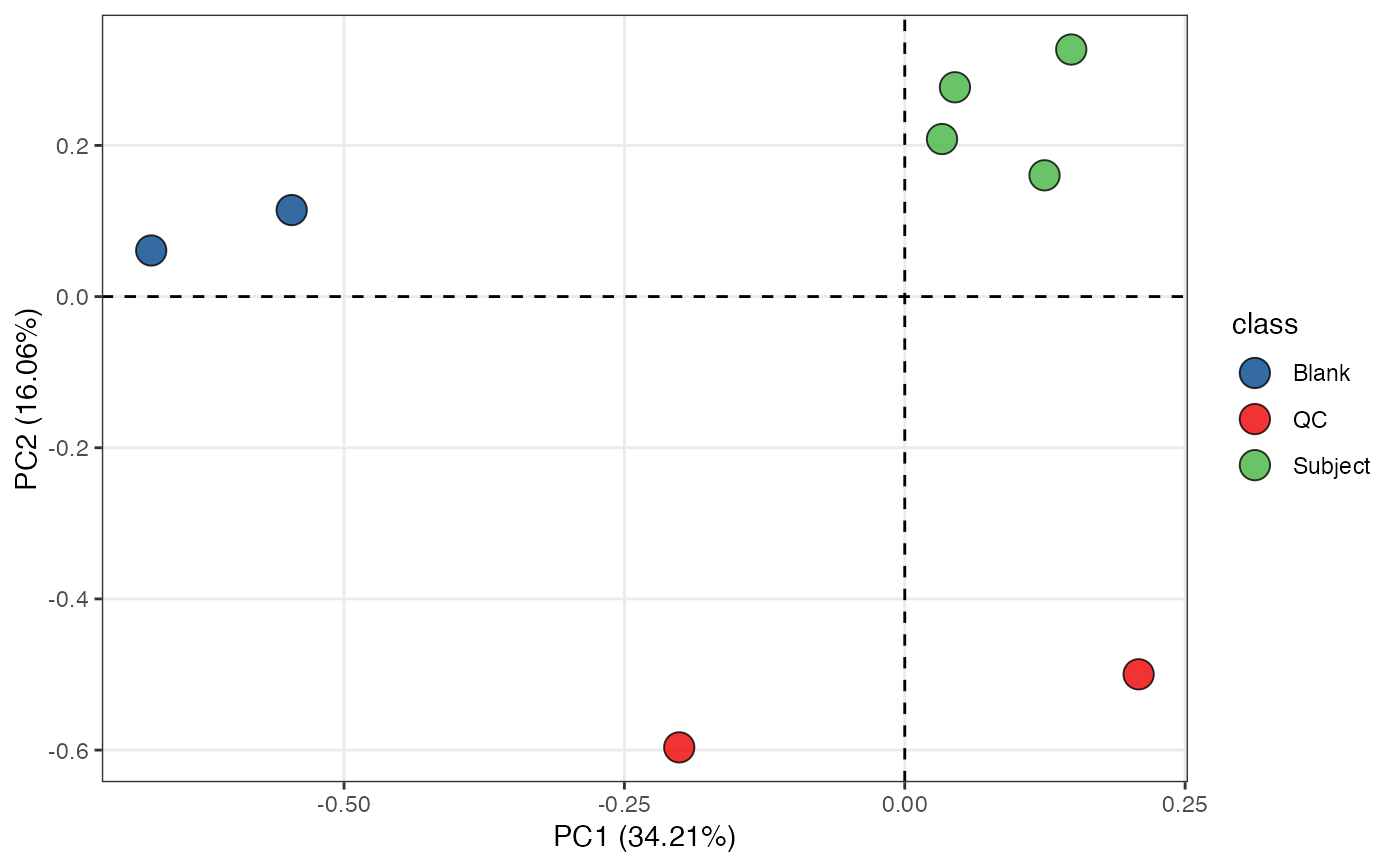 object %>%
scale %>%
massqc_pca(color_by = "class", frame = FALSE) +
ggsci::scale_fill_lancet() +
ggrepel::geom_text_repel(aes(label = sample_id))
#> Warning: MVs in you object,
#>
#> will remove variables > 50% and imputate with zero.
object %>%
scale %>%
massqc_pca(color_by = "class", frame = FALSE) +
ggsci::scale_fill_lancet() +
ggrepel::geom_text_repel(aes(label = sample_id))
#> Warning: MVs in you object,
#>
#> will remove variables > 50% and imputate with zero.
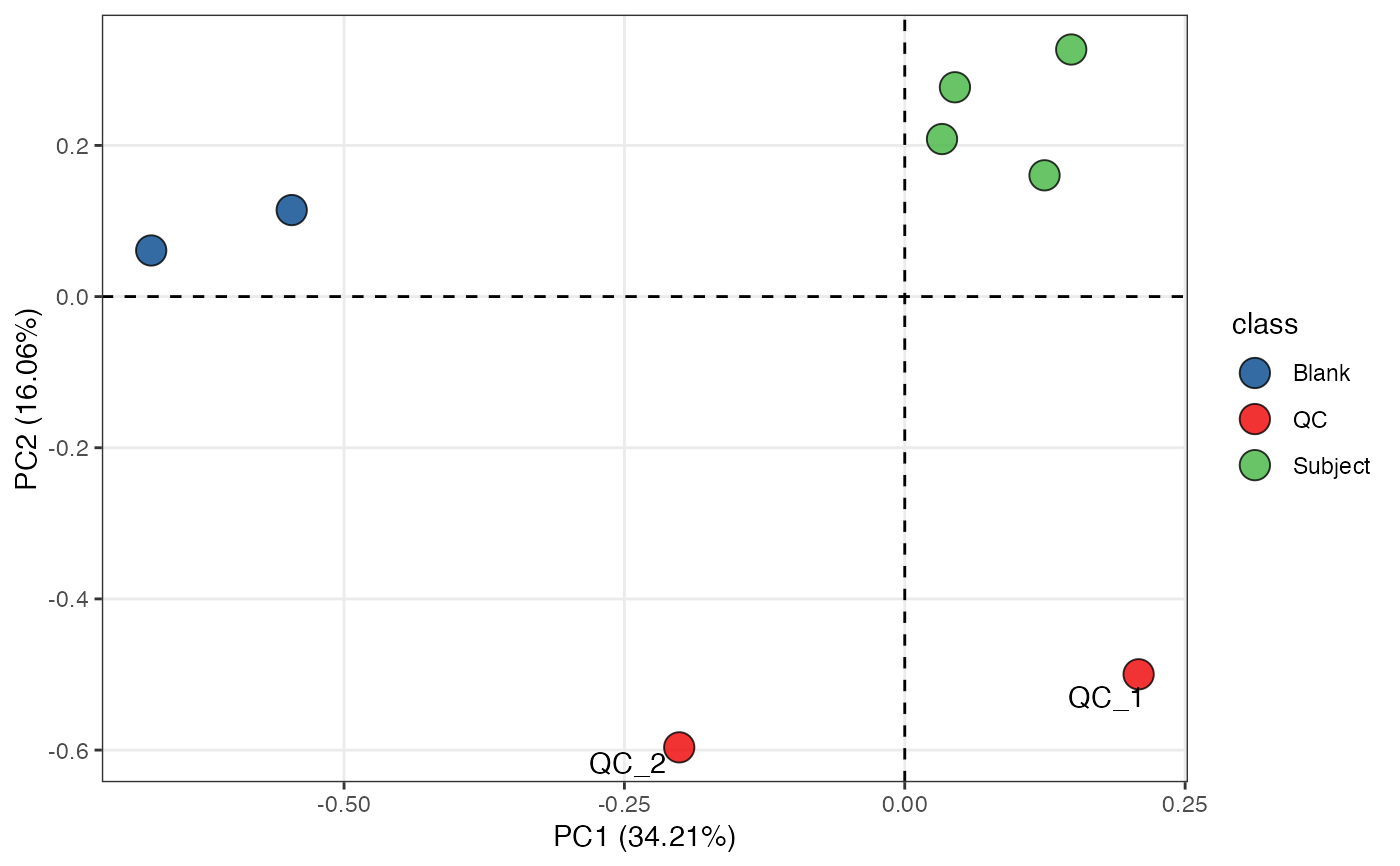
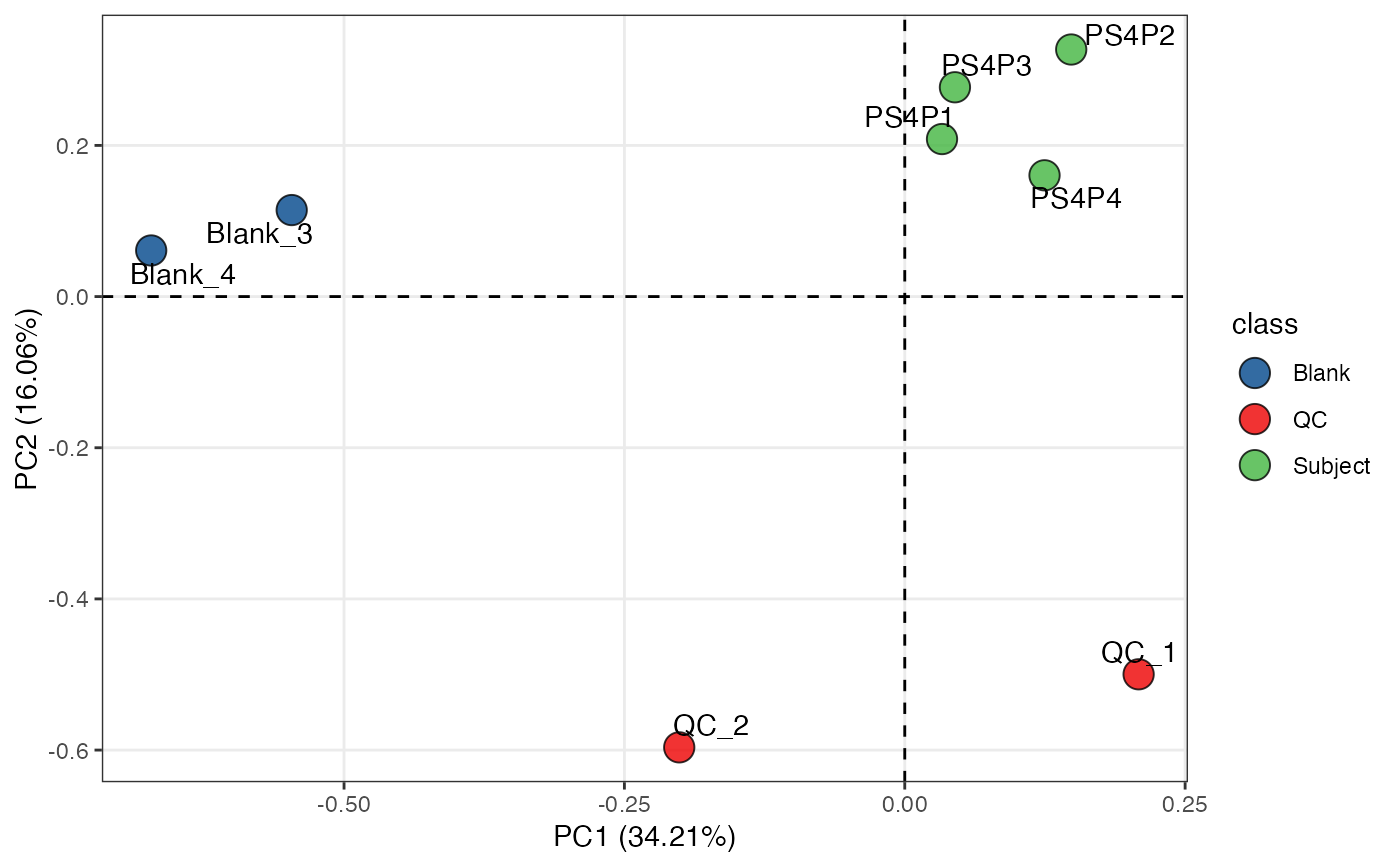 object %>%
scale %>%
massqc_pca(color_by = "class", frame = FALSE) +
ggsci::scale_fill_lancet() +
ggrepel::geom_text_repel(aes(label = ifelse(class == "QC", sample_id, NA)))
#> Warning: MVs in you object,
#>
#> will remove variables > 50% and imputate with zero.
#> Warning: Removed 6 rows containing missing values (geom_text_repel).
object %>%
scale %>%
massqc_pca(color_by = "class", frame = FALSE) +
ggsci::scale_fill_lancet() +
ggrepel::geom_text_repel(aes(label = ifelse(class == "QC", sample_id, NA)))
#> Warning: MVs in you object,
#>
#> will remove variables > 50% and imputate with zero.
#> Warning: Removed 6 rows containing missing values (geom_text_repel).